____________________
Databases:

M2OR (from M. Lalis, et al. Nucleic. Acid. Res. 2024) A Database of Olfactory Receptor-Odorant Pairs for Understanding the Molecular Mechanisms of Olfaction

SweetenersDB v2.0 (from C. Bouysset et al. Food Chem. 2020) a database of sweeteners with known sweetness values
SweetenersDB v1.0 (from J.B. Chéron et al. Food Chem. 2016)
____________________
Softwares and webservers:
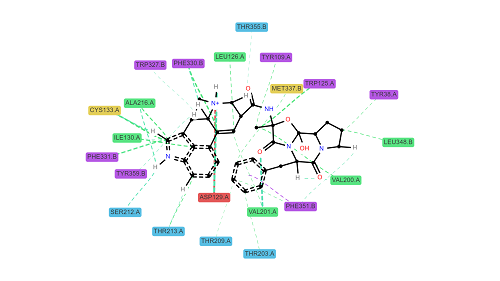
ProLIF (Protein-Ligand Interaction Fingerprints), a tool designed to generate interaction fingerprints for protein-ligand and protein-protein interactions extracted from molecular dynamics trajectories and docking simulations. It also supports DNA-ligand, DNA-protein and DNA-DNA interactions. (Github)

Predisweet, a webserver for sweet taste prediction (webserver)
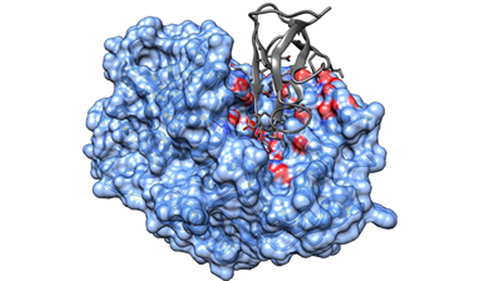
Attract FF2G for protein-protein binding affinity prediction (webserver)
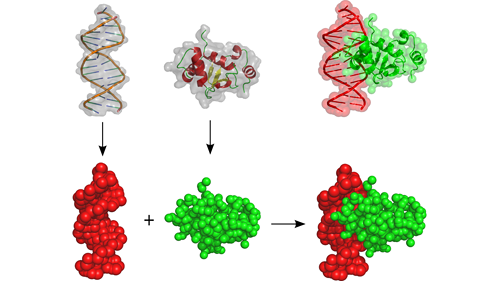
Ptools source code (ptools_1.1.tar.gz released in 2015-03-15) including the following features:
– Attract forcefields FF1 and FF2 for proteins and nucleic acids
– Attract docking protocol and multidocking script for 3-body systems
– Analysis scripts (RMSd, iRMSd, fnat, clustering, …)
– Prediction of protein interface based on electrostatic desolvation scoring
Ptools/Attract (source and binaries): Original Protein-Protein Docking software

