The aim of our research is to decipher structure-function relationships of mammals and insect proteins involved in olfaction and taste, such as Olfactory Receptor (OR), Taste Receptor (TASR) or Odorant Binding Protein (OBP) combining computational approaches (molecular dynamics simulations, virtual screening and structural bioinformatics) and in vitro experiments.
Latest published works:
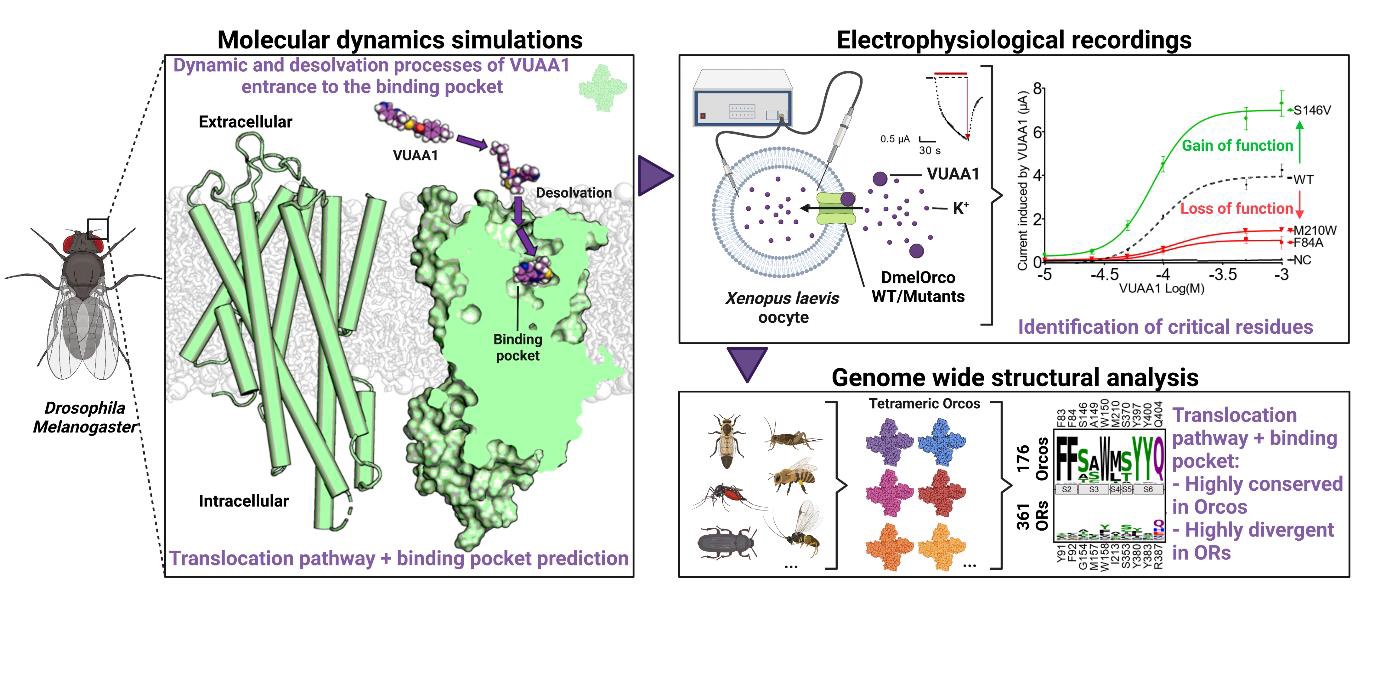
Elucidation of the structural basis for ligand binding and translocation in conserved insect odorant receptor co-receptors
Abstract: In numerous insects, the olfactory receptor family forms a unique class of heteromeric cation channels. Recent progress in resolving the odorant receptor structures offers unprecedented opportunities for deciphering their molecular mechanisms of ligand recognition. Unexpectedly, these structures in apo or ligand-bound states did not reveal the pathway taken by the ligands between the extracellular space and the deep internal cavities. By combining molecular modeling with electrophysiological recordings, we identified amino acids involved in the dynamic entry pathway and the binding of VUAA1 to Drosophila melanogaster’s odorant receptor co-receptor (Orco). Our results provide evidence for the exact location of the agonist binding site and a detailed and original mechanism of ligand translocation controlled by a network of conserved residues. These findings would explain the particularly high selectivity of Orcos for their ligands.
J. Pacalon, G. Audic, J. Magnat, M. Philip, J. Golebiowski, C.J. Moreau, J. Topin, Nat. Commun. 2023, 14, 8182. DOI
Ligand Binding Properties of Odorant-Binding Protein OBP5 from Mus musculus
Abstract: Odorant-binding proteins (OBPs) are abundant soluble proteins secreted in the nasal mucus of a variety of species that are believed to be involved in the transport of odorants toward olfactory receptors. In this study, we report the functional characterization of mouse OBP5 (mOBP5). mOBP5 was recombinantly expressed as a hexahistidine-tagged protein in bacteria and purified using metal affinity chromatography. The oligomeric state and secondary structure composition of mOBP5 were investigated using gel filtration and circular dichroism spectroscopy. Fluorescent experiments revealed that mOBP5 interacts with the fluorescent probe N-phenyl naphthylamine (NPN) with micromolar affinity. Competitive binding experiments with 40 odorants indicated that mOBP5 binds a restricted number of odorants with good affinity. Isothermal titration calorimetry (ITC) confirmed that mOBP5 binds these compounds with association constants in the low micromolar range. Finally, protein homology modeling and molecular docking analysis indicated the amino acid residues of mOBP5 that determine its binding properties.
L. Moitrier, C. Belloir, M. Lalis, Y. Hou, J. Topin, L. Briand, Biol. 2022, 12, 2. DOI
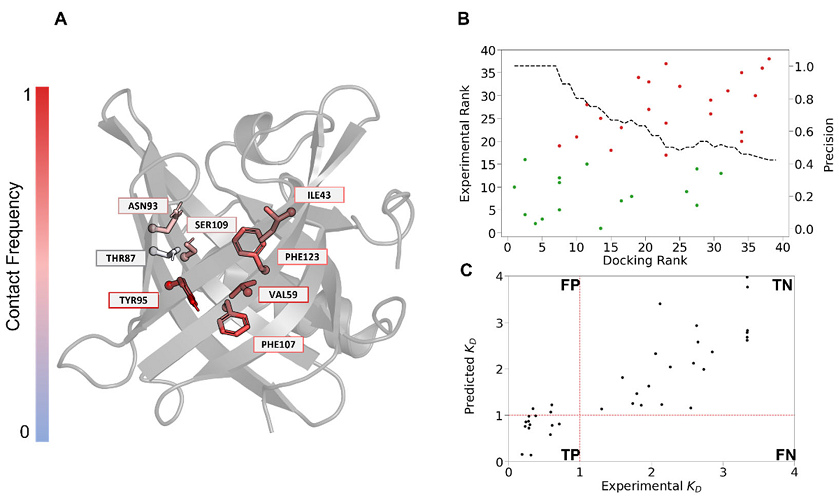
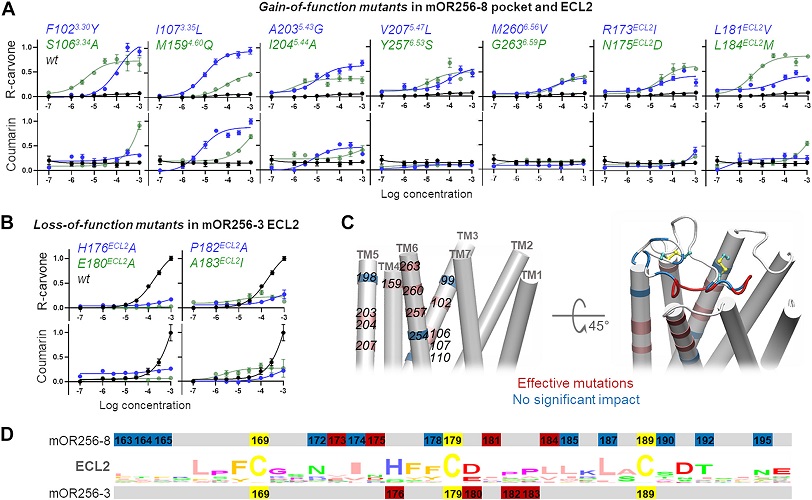
Extracellular loop 2 of G protein–coupled olfactory receptors is critical for odorant recognition
Abstract: G protein–coupled olfactory receptors (ORs) enable us to detect innumerous odorants. They are also ectopically expressed in nonolfactory tissues and emerging as attractive drug targets. ORs can be promiscuous or highly specific, which is part of a larger mechanism for odor discrimination. Here, we demonstrate that the OR extracellular loop 2 (ECL2) plays critical roles in OR promiscuity and specificity. Using site-directed mutagenesis and molecular modeling, we constructed 3D OR models in which ECL2 forms a lid over the orthosteric pocket. We demonstrate using molecular dynamics simulations that ECL2 controls the shape and volume of the odorant-binding pocket, maintains the pocket hydrophobicity, and acts as a gatekeeper of odorant binding. Therefore, we propose the interplay between the specific orthosteric pocket and the variable, less specific ECL2 controls OR specificity and promiscuity. Furthermore, the 3D models created here enabled virtual screening of new OR agonists and antagonists, which exhibited a 70% hit rate in cell assays. Our approach can potentially be generalized to structure-based ligand screening for other G protein–coupled receptors that lack high-resolution 3D structures.
Y. Yu, Z. Ma, J. Pacalon, L. Xu, W. Li, C. Belloir, J. Topin, L. Briand, J. Golebiowski, X. Cong, J. Biol. Chem. 2022, 298(9), 102331. DOI
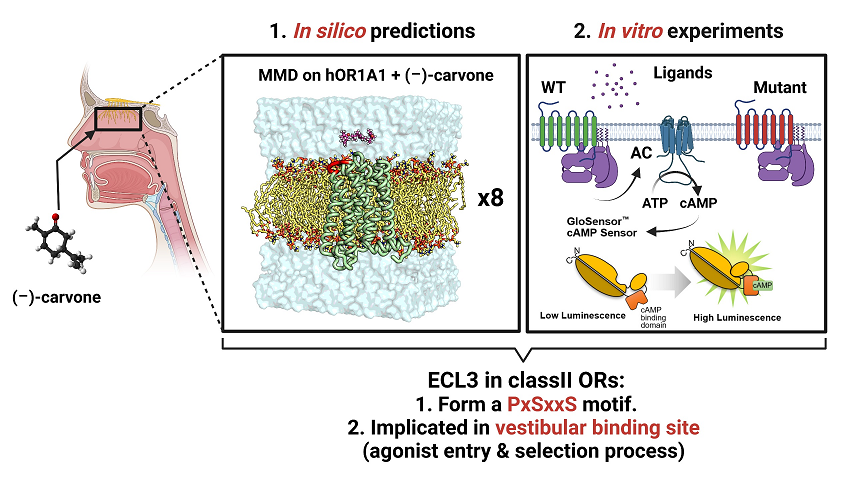
The Third Extracellular Loop of Mammalian Odorant Receptors Is Involved in Ligand Binding
Abstract: Mammals recognize chemicals in the air via G protein-coupled odorant receptors (ORs). In addition to their orthosteric binding site, other segments of these receptors modulate ligand recognition. Focusing on human hOR1A1, which is considered prototypical of class II ORs, we used a combination of molecular modeling, site-directed mutagenesis, and in vitro functional assays. We showed that the third extracellular loop of ORs (ECL3) contributes to ligand recognition and receptor activation. Indeed, site-directed mutations in ECL3 showed differential effects on the potency and efficacy of both carvones, citronellol, and 2-nonanone.
T. Shim, J. Pacalon, W. C. Kim, X. Cong, J. Topin, J. Golebiowski, , & C. Moon, Int. J. Mol. Sci. 2022, 23(20), 12501. DOI
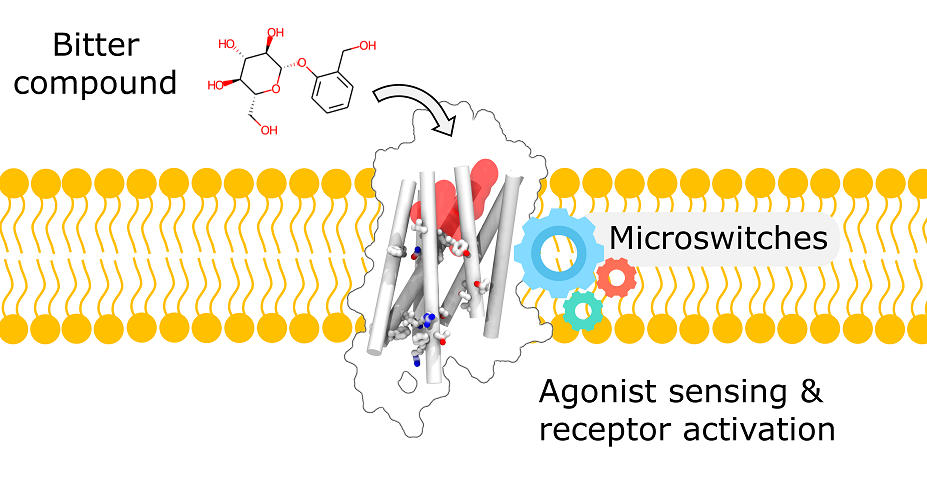
Functional Molecular Switches of Mammalian G Protein-Coupled Bitter-Taste Receptors.
Abstract: Bitter taste receptors (TAS2Rs) are a poorly understood subgroup of G protein-coupled receptors (GPCRs). The experimental structure of these receptors has yet to be determined, and key-residues controlling their function remain mostly unknown. We designed an integrative approach to improve comparative modeling of TAS2Rs. Using current knowledge on class A GPCRs and existing experimental data in the literature as constraints, we pinpointed conserved motifs to entirely re-align the amino-acid sequences of TAS2Rs. We constructed accurate homology models of human TAS2Rs. As a test case, we examined the accuracy of the TAS2R16 model with site-directed mutagenesis and in vitro functional assays. This combination of in silico and in vitro results clarifies sequence-function relationships and proposes functional molecular switches that encode agonist sensing and downstream signaling mechanisms within mammalian TAS2Rs sequences.
J. Topin, C. Bouysset, J. Pacalon, Y. Kim, M. Rhyu, S. Fiorucci, J. Golebiowski. Cell. Mol. Life Sci. 2021, 78, 7605-7615 DOI
