Our brain perceives our molecular environment through two chemical senses, olfaction and taste, by triggering the activation of chemical sensors named olfactory and gustatory receptors. These receptors are differentially activated by a virtually infinite number of molecules suggesting a extremely complex combinatorial code. Can a computer learn how to smell and taste? The objective of our research, involving chemists, computer scientists and neurobiologists, is to push the boundaries of artificial intelligence models in the context of chemical senses.
Latest published works:
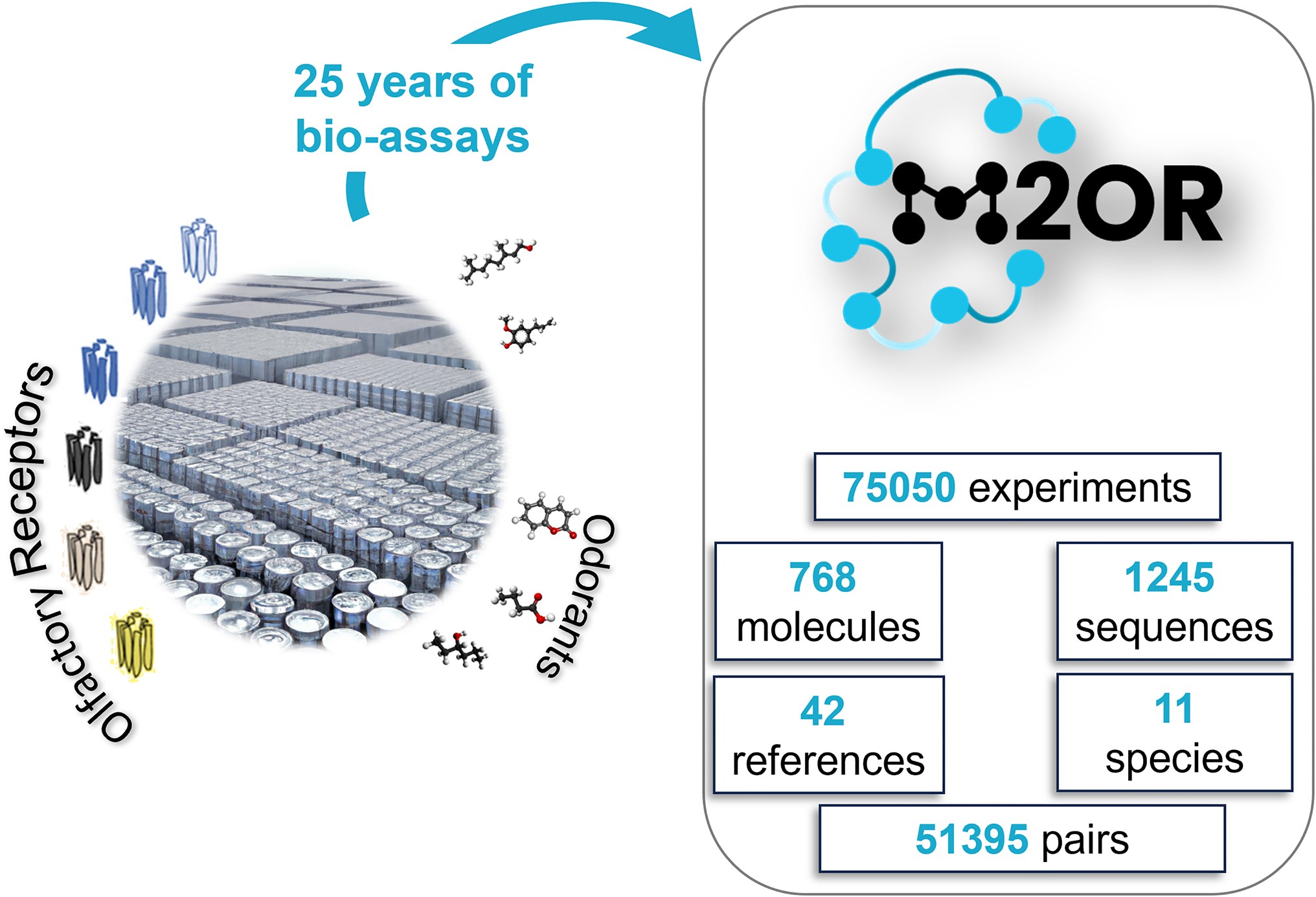
M2OR: A Database of Olfactory Receptor-Odorant Pairs for Understanding the Molecular Mechanisms of Olfaction
Abstract: Mammalian sense of smell is triggered by interaction between odorant molecules and a class of proteins, called olfactory receptors (ORs). These receptors, expressed at the surface of olfactory sensory neurons, encode myriad of distinct odors via a sophisticated activation pattern. However, determining the molecular recognition spectrum of ORs remains a major challenge. The Molecule to Olfactory Receptor database (M2OR, https://m2or.chemsensim.fr/) provides curated data that allows an easy exploration of the current state of the research on OR-molecule interaction. We have gathered a database of 75,050 bioassay experiments for 51 395 distinct OR-molecule pairs. Drawn from published literature and public databases, M2OR contains information about OR responses to molecules and their mixtures, receptor sequences and experimental details. Users can obtain information on the activity of a chosen molecule or a group of molecules, or search for agonists for a specific OR or a group of ORs. Advanced search allows for fine-grained queries using various metadata such as species or experimental assay system, and the database can be queried by multiple inputs via a batch search. Finally, for a given search query, users can access and download a curated aggregation of the experimental data into a binarized combinatorial code of olfaction.
M. Lalis, M. Hladiš, S. Abi Khalil, S. Fiorucci, L. Briand, J. Topin. Nucleic. Acids. Res. 2024, 52, D1370–D1379. DOI
Matching receptor to odorant with protein language and graph neural networks
Abstract: Odor perception in mammals is triggered by interactions between volatile organic compounds and a subset of hundreds of proteins called olfactory receptors (ORs). Molecules activate these receptors in a complex combinatorial coding allowing mammals to discriminate a vast number of chemical stimuli. Recently, ORs have gained attention as new therapeutic targets following the discovery of their involvement in other physiological processes and diseases. To date, predicting molecule-induced activation for ORs is highly challenging since 43% of ORs have no identified active compound. In this work, we combine [CLS] token from protBERT with a molecular graph and propose a tailored GNN architecture incorporating inductive biases from the protein-molecule binding. We abstract the biological process of protein-molecule activation as the injection of a molecule into a protein-specific environment. On a newly gathered dataset of 46 700 OR-molecule pairs, this model outperforms state-of-the-art models on drug-target interaction prediction as well as standard GNN baselines. Moreover, by incorporating non-bonded interactions the model is able to work with mixtures of compounds. Finally, our predictions reveal a similar activation pattern for molecules within a given odor family, which is in agreement with the theory of combinatorial coding in olfaction
M. Hladiš, M. Lalis, S. Fiorucci, J. Topin. International Conference on Learning Representation (ICLR), 2023, DOI (OpenReview)
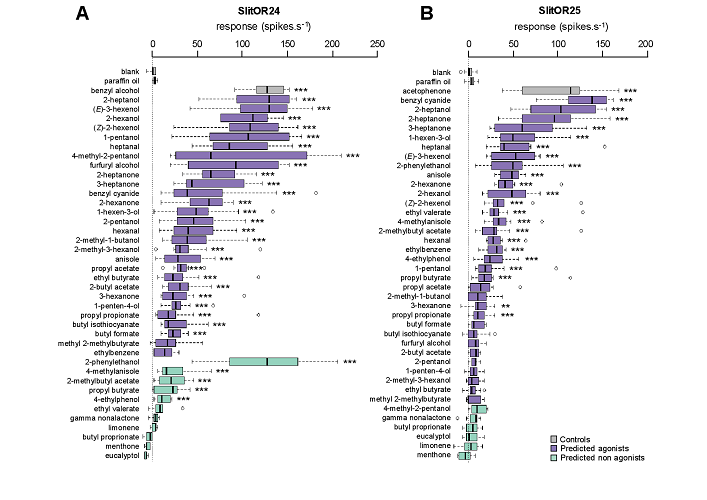
Reverse chemical ecology in a moth: machine learning on odorant receptors identifies new behaviorally active agonists
Abstract: The concept of reverse chemical ecology (exploitation of molecular knowledge for chemical ecology) has recently emerged in conservation biology and human health. Here, we extend this concept to crop protection. Targeting odorant receptors from a crop pest insect, the noctuid moth Spodoptera littoralis, we demonstrate that reverse chemical ecology has the potential to accelerate the discovery of novel crop pest insect attractants and repellents. Using machine learning, we first predicted novel natural ligands for two odorant receptors, SlitOR24 and 25. Then, electrophysiological validation proved in silico predictions to be highly sensitive, as 93% and 67% of predicted agonists triggered a response in Drosophila olfactory neurons expressing SlitOR24 and SlitOR25, respectively, despite a lack of specificity. Last, when tested in Y-maze behavioral assays, the most active novel ligands of the receptors were attractive to caterpillars. This work provides a template for rational design of new eco-friendly semiochemicals to manage crop pest populations.
G. Caballero-Vidal, C. Bouysset, J. Gévar, H. Mbouzid, C. Nara, J. Delaroche, J. Golebiowski, N. Montagné, S. Fiorucci, E. Jacquin-Joly. Cell. Mol. Life Sci. 2021, 78, 6593-6603 DOI
Machine learning decodes chemical features to identify novel agonists of a moth odorant receptor
Abstract: Odorant receptors expressed at the peripheral olfactory organs are key proteins for animal volatile sensing. Although they determine the odor space of a given species, their functional characterization is a long process and remains limited. To date, machine learning virtual screening has been used to predict new ligands for such receptors in both mammals and insects, using chemical features of known ligands. In insects, such approach is yet limited to Diptera, whereas insect odorant receptors are known to be highly divergent between orders. Here, we extend this strategy to a Lepidoptera receptor, SlitOR25, involved in the recognition of attractive odorants in the crop pest Spodoptera littoralis larvae. Virtual screening of 3 million molecules predicted 32 purchasable ones whose function has been systematically tested on SlitOR25, revealing 11 novel agonists with a success rate of 28%. Our results show that Support Vector Machine optimizes the discovery of novel agonists and expands the chemical space of a Lepidoptera OR. More, it opens up structure-function relationship analyses through a comparison of the agonist chemical structures. This proof-of-concept in a crop pest could ultimately enable the identification of OR agonists or antagonists, capable of modifying olfactory behaviors in a context of biocontrol.
G. Caballero-Vidal, C. Bouysset, H. Grunig, S. Fiorucci, N. Montagné, J. Golebiowski, E. Jacquin-Joly. Sci. Rep. 2020, 1655. DOI


Novel scaffold of natural compound eliciting sweet taste revealed by machine learning
Abstract: Sugar replacement is still an active issue in the food industry. The use of structure-taste relationships remains one of the most rational strategy to expand the chemical space associated to sweet taste. A new machine learning model has been setup based on an update of the SweetenersDB and on open-source molecular features. It has been implemented on a freely accessible webserver. Cellular functional assays show that the sweet taste receptor is activated in vitro by a new scaffold of natural compounds identified by the in silico protocol. The newly identified sweetener belongs to the lignan chemical family and opens a new chemical space to explore.
C. Bouysset, C. Belloir, S. Antonczak, L. Briand, S. Fiorucci. Food Chem. 2020, 324, 126864. DOI
